Introduction: Unlocking the Secrets of Gene Regulation
In the ever-evolving world of genomics and bioinformatics, understanding the intricate mechanisms of gene expression is crucial. One of the key elements in this process is the cis-regulatory element non-coding DNA sequences that regulate the transcription of neighboring genes.
This is where cisRED comes in.
cisRED (cis-Regulatory Element Database) is a robust, genome-wide analytical platform built to help researchers uncover conserved regulatory motifs across species. By leveraging evolutionary conservation and gene co-expression data, cisRED delivers a curated set of high-confidence regulatory elements for the scientific community.
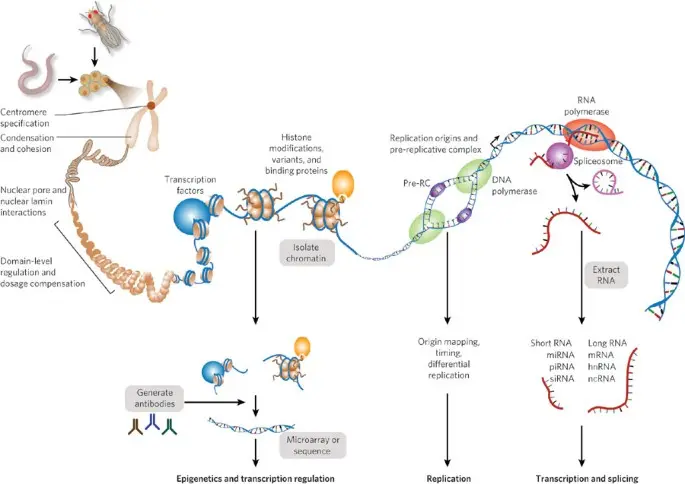
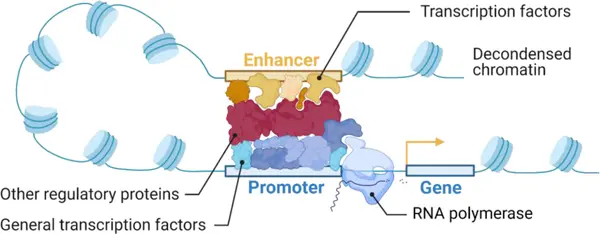
cisRED is a fully automated system designed to identify regulatory sequences in the non-coding regions upstream of genes. These upstream regions are particularly important because they often contain transcription factor binding sites (TFBS) the control switches that regulate whether a gene is turned on or off.
To ensure accuracy and relevance, cisRED focuses on:
- 1.5 kb upstream of each Transcription Start Site (TSS)
- Filtering out repetitive elements
- Detecting statistically significant motifs
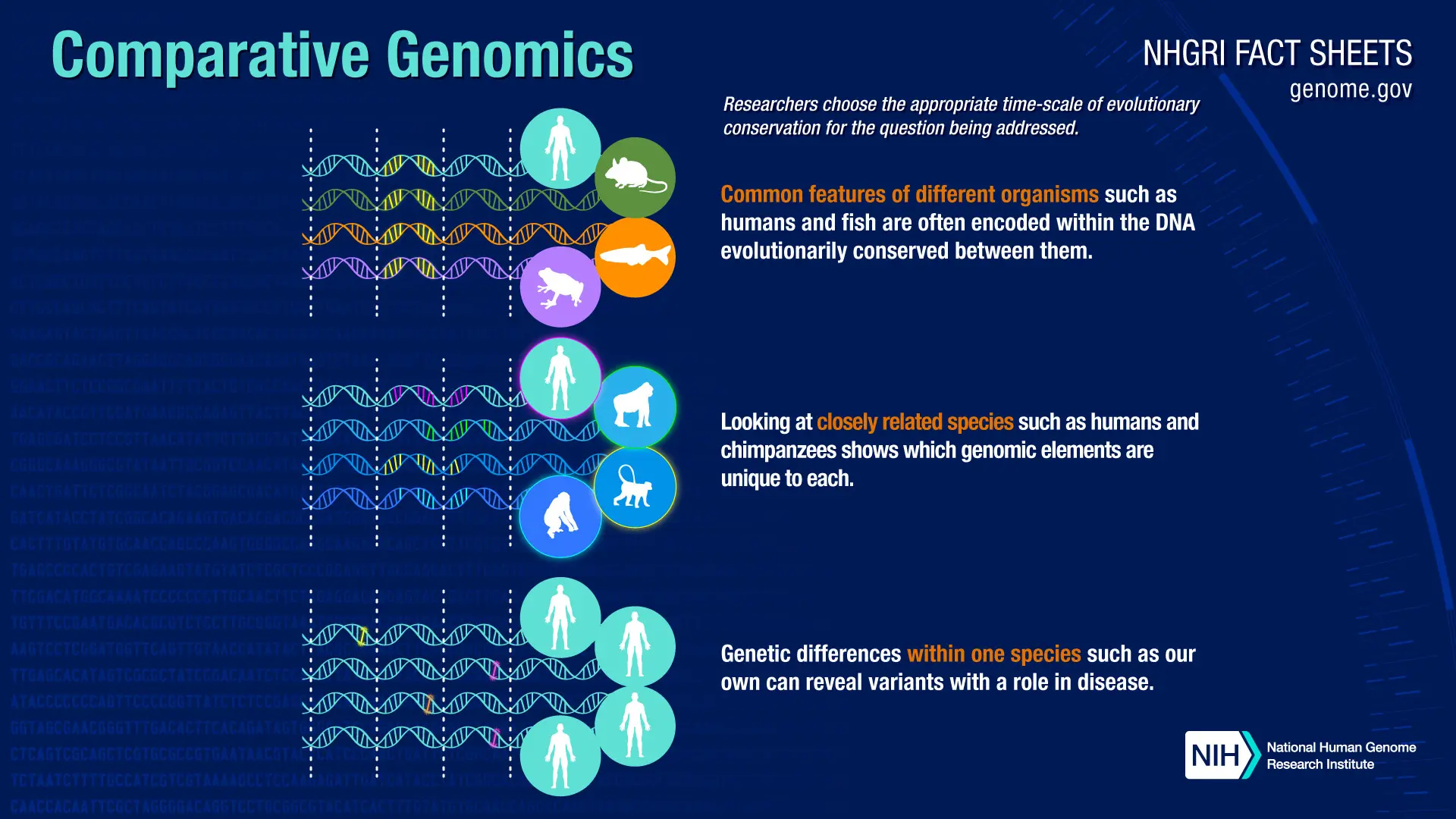
- Using comparative genomics to analyze orthologous genes across multiple species
This approach allows cisRED to offer computationally predicted regulatory motifs that are conserved and biologically relevant.
Whether you're working in molecular biology, genomics, systems biology, or computational biology, cisRED provides tools that are highly valuable for:
cisRED helps scientists identify transcription factor binding sites involved in cell-specific or condition-specific gene expression.
Comparative Genomics
By comparing conserved motifs across species, cisRED supports the study of evolutionarily conserved regulatory mechanisms.
Bioinformatics Tool Development
Developers can integrate cisRED data into new tools or pipelines aimed at motif analysis, gene network prediction, or regulatory sequence annotation.
Functional Genomics
cisRED helps experimentalists prioritize candidate regulatory regions for further testing in reporter assays, ChIP-Seq, or CRISPR screens.
Here’s a simplified workflow of the cisRED platform:
- Genomic sequences from well-annotated species
- Co-expressed gene sets and orthologous gene alignments
1.Data Collection
- Use of multiple algorithms to scan upstream regions
- Focus on non-repetitive, conserved segments
2. Motif Discovery
3.Statistical Filtering
- Remove low-confidence or non-informative motifs
- Retain motifs with functional relevance
4.Database Curation and Access
- Public access via user-friendly interface
-
Tools for motif visualization, gene query, and cross-species comparison
- Genome-Wide Motif Analysis
- Conserved Motif Identification
- Focus on Functional Non-Coding Regions
- Cross-Species Comparisons
- Statistical Significance Filters
- Biologically Informed Algorithms
cisRED isn't just a tool it's a community-driven platform aimed at accelerating discoveries in gene regulation. As researchers continue to unravel the complexities of the genome, resources like cisRED become indispensable.
By combining computational predictions with biological insights, cisRED bridges the gap between in silico and in vivo research, enabling more targeted and effective scientific investigations.
Understanding the regulatory logic of the genome is essential for advances in medicine, biotechnology, and genetic research. cisRED equips scientists with powerful data and tools to decode the hidden language of gene control.
Ready to dive into the world of cis-regulatory elements?
Explore cisRED today and bring your genomic research to the next level.