In the vast landscape of the genome, only a small fraction about 1–2% codes for proteins. For decades, scientists focused their attention on these protein-coding genes, believing they were the primary actors in biology. But as genomic research evolved, a new paradigm emerged: the real control lies outside the genes, in the non-coding regions that regulate when and where genes are expressed.
This blog explores how DNA motifs and cis-regulatory elements orchestrate gene expression, and how tools like cisRED are helping researchers uncover this hidden regulatory code.
Gene regulation refers to the mechanisms that control the activity of genes specifically, how much of a gene’s product (RNA or protein) is made, and when or where it’s produced. This process ensures that:
- The right genes are expressed in the right cells
- Genes are activated in response to environmental signals
- Cells differentiate into distinct types (e.g., muscle, nerve, liver)
Without regulation, all cells would express all genes all the time which would be chaotic and biologically unsustainable.
➤ What Are DNA Motifs?
DNA motifs are short, recurring sequences (typically 6–15 base pairs) that serve as binding sites for transcription factors (TFs). These motifs are not random they are conserved, meaning they appear consistently across individuals or species, suggesting functional importance.
➤ What Are Cis-Regulatory Elements?
Cis-regulatory elements (CREs) are non-coding regions of DNA that contain these motifs. They include:
- Promoters : located just upstream of a gene; essential for transcription initiation
- Enhancers : can be located far from the gene; boost transcription levels
- Silencers : suppress gene expression
- Insulators : prevent interactions between unrelated genes
Together, CREs act like switches and dials, fine-tuning gene expression in space and time.
The discovery of DNA motifs has been revolutionized by bioinformatics and genomic databases. Traditional lab methods like ChIP-seq (Chromatin Immunoprecipitation Sequencing) are now paired with computational tools that scan thousands of sequences for statistically significant patterns.
Tools like cisRED apply genome-wide searches for:
- Conserved motifs across species (orthologs)
- Motif enrichment in co-expressed genes
- Position-specific scoring to estimate binding probabilities
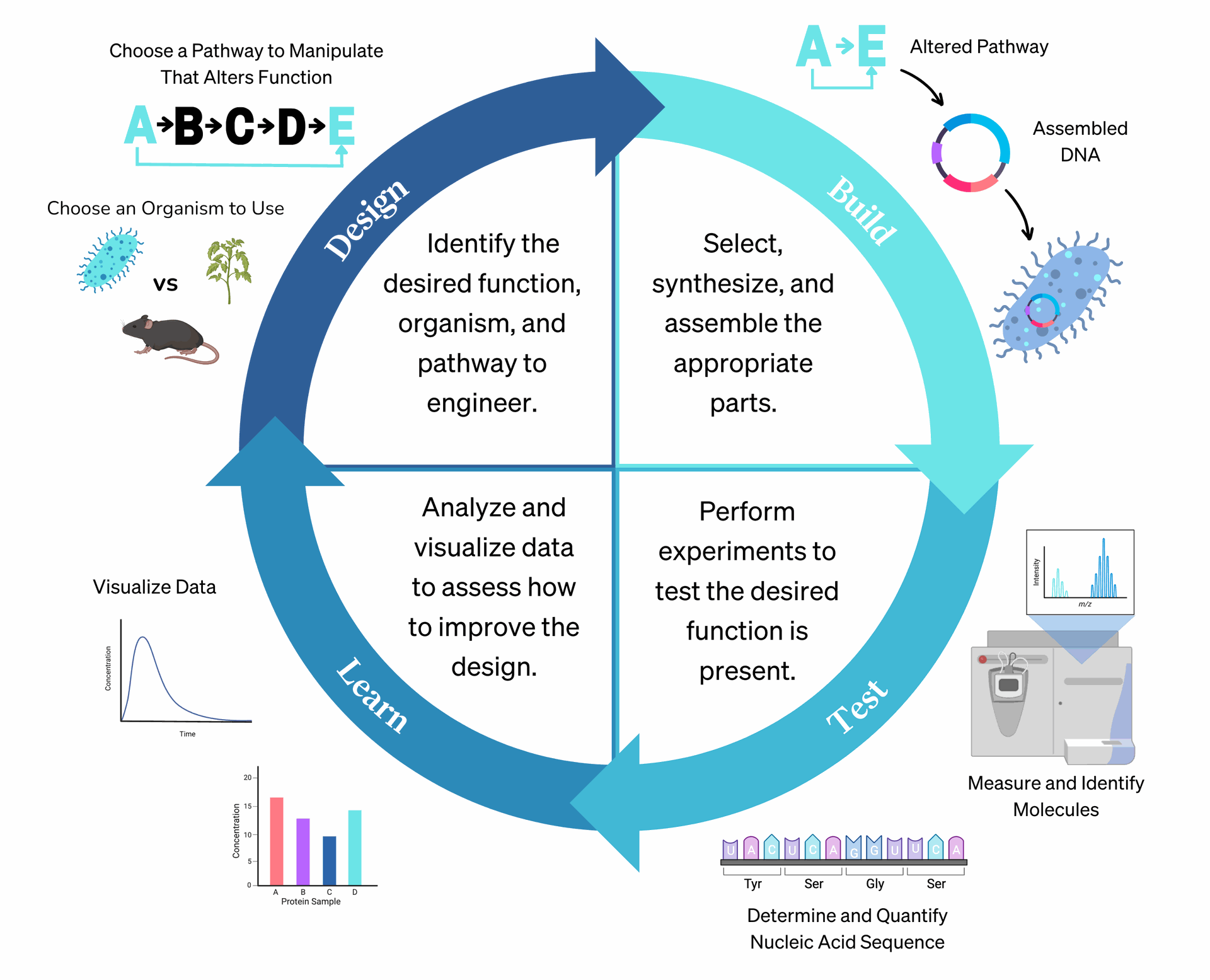
Motif discovery involves:
- Selecting a target gene
- Extracting upstream sequences (e.g., 1.5 kb before the TSS)
- Comparing with co-expressed genes and orthologues
- Detecting over-represented sequences
- Generating Position Weight Matrices (PWMs) and logos
🔹 Disease Research
Many disease-causing mutations don’t affect genes directly they affect regulatory motifs, disrupting transcription factor binding. Examples:
- Cancer: Mutations in p53 binding sites
- Autoimmune disorders: Altered enhancers in immune genes
- Neurodevelopmental disorders: Changes in brain-specific enhancers
🔹 Development and Cell Identity
Regulatory motifs are responsible for activating developmental genes at the right time and place, defining cell fate and organ formation.
🔹 Evolutionary Conservation
Motifs that are conserved across species are likely to be functionally essential. Comparative genomics reveals which motifs are preserved and under evolutionary pressure.
The cisRED database (cis-Regulatory Element Discovery) is an automated system that identifies and ranks conserved regulatory motifs across multiple genomes. It focuses on 1.5 kb upstream regions, net of repetitive elements.www.cisred.org
Key features:
- Supports multiple organisms (e.g., Human, Mouse, C. elegans)
- Detects motifs using co-expression and orthology data
-
Provides results as:
- Position Frequency Matrices (PFMs)
- Hidden Markov Models (HMMs)
- Sequence logos (JPG)
-
Offers data via:
- Web interface
- Remote MySQL
- Bulk FTP downloads
cisRED helps scientists answer questions like:
- What transcription factors might regulate a gene?
- Which motifs are conserved across species?
- What patterns appear in co-expressed gene sets?
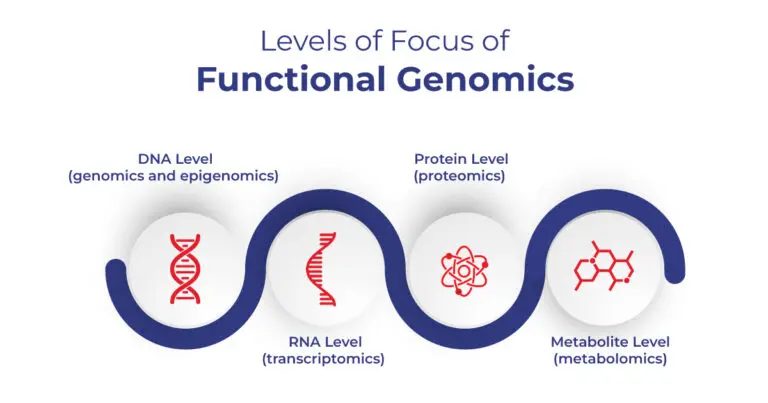
- Functional Genomics: Predicting which DNA sequences are biologically active
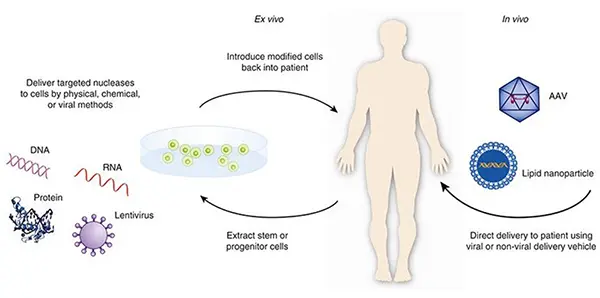
- Gene Therapy: Avoiding or targeting regulatory regions during editing
- Synthetic Biology: Designing artificial promoters and enhancers
- Drug Discovery: Targeting regulatory pathways for treatment