Understanding Cis-Regulatory Elements and How cisRED Drives Breakthroughs in Gene Regulation
Understanding gene regulation is fundamental to unlocking the mysteries of biology, disease mechanisms, and therapeutic development. At the core of gene regulation are cis-regulatory elements DNA sequences such as promoters and enhancers that control when and where genes are expressed. Accurately identifying these elements is essential for researchers in genomics, molecular biology, and bioinformatics.
This is where cisRED comes in: a comprehensive, genome-wide database and analytical platform designed to discover and predict conserved cis-regulatory motifs across species. By providing high-confidence regulatory sequences, cisRED enables researchers to deepen their understanding of transcriptional control and accelerate their research.
In this blog, we’ll explore how cisRED empowers researchers, discuss the underlying techniques, address common challenges, and highlight how the right experimental tools and reagents can amplify your discovery process helping you translate computational insights into robust biological validation.
What Is cisRED and Why Is It Important?
cisRED is a comprehensive, genome-wide platform that helps scientists identify conserved DNA sequences involved in gene regulation. Specifically, it focuses on regions located about 1.5 kilobases upstream of transcription start sites (TSS), where transcription factor binding sites (TFBS) key regulators of gene expression are commonly found.
By analyzing thousands of genes across multiple species, cisRED leverages evolutionary conservation to highlight regulatory motifs that are most likely to play important roles in controlling gene activity. The platform applies advanced computational algorithms combined with stringent statistical filters to eliminate repetitive or irrelevant sequences, resulting in a reliable collection of candidate regulatory elements.
Researchers in genomics, bioinformatics, molecular biology, and related fields benefit from cisRED’s user-friendly interface that allows easy querying, visualization, and download of regulatory motifs. This empowers scientists to formulate hypotheses about gene regulation mechanisms, select promising motifs for experimental validation, and accelerate discoveries in areas ranging from basic biology to disease research.
Key Techniques Behind cisRED
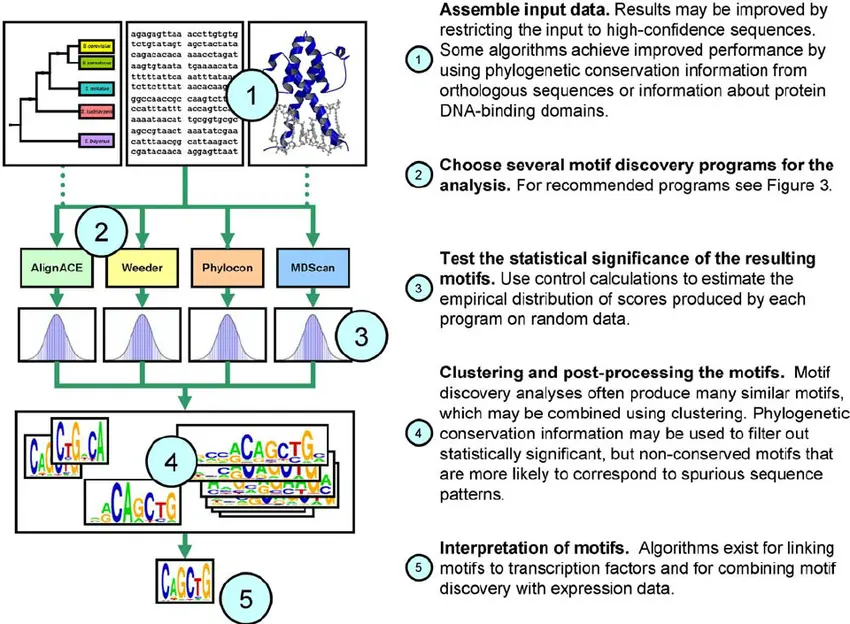
Motif Discovery Algorithms
cisRED applies multiple computational algorithms to detect short DNA sequences (motifs) that occur more frequently in conserved non-coding regions than expected by chance. These include:
- Phylogenetic footprinting: Comparing orthologous sequences to find conserved motifs.
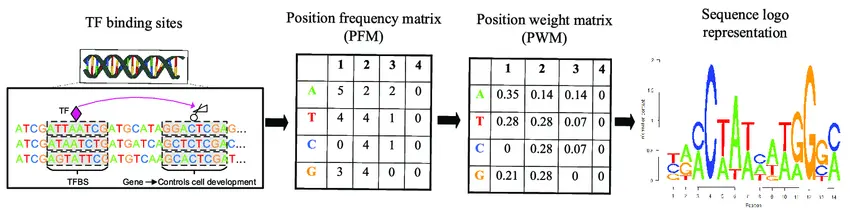
- Position weight matrices (PWMs): Modeling motif variability to capture binding site flexibility.
- Statistical enrichment analysis: Differentiating biologically relevant motifs from background noise.
Filtering and Annotation
Post-discovery, motifs undergo filtering to exclude repetitive elements and low-confidence hits. Annotated motifs are cross-referenced with known transcription factor binding databases to provide functional insights.
Common Challenges in Motif Discovery and How to Overcome Them
1. Distinguishing True Motifs from Noise
Motif discovery can produce many false positives. Using conserved regions across species, as cisRED does, greatly improves specificity.
2. Handling Repetitive Elements
Repetitive sequences can mimic motif patterns. cisRED’s pipeline excludes repetitive elements to enhance result quality.
3. Translating Computational Predictions to Biological Validation
Computational motifs need experimental validation a critical step where specialized reagents and tools come into play (see next section).
From Prediction to Validation: Tools and Reagents That Enhance Your Research
Predicting cis-regulatory motifs is only half the journey. To confirm that predicted motifs functionally regulate gene expression, experimental approaches such as:
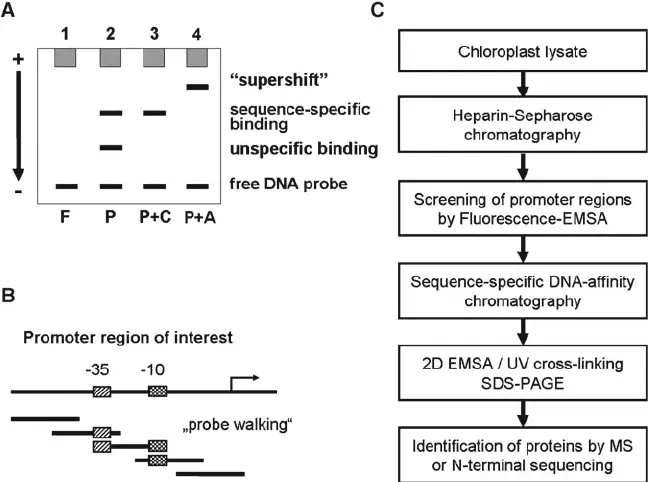
- Electrophoretic mobility shift assays (EMSA)
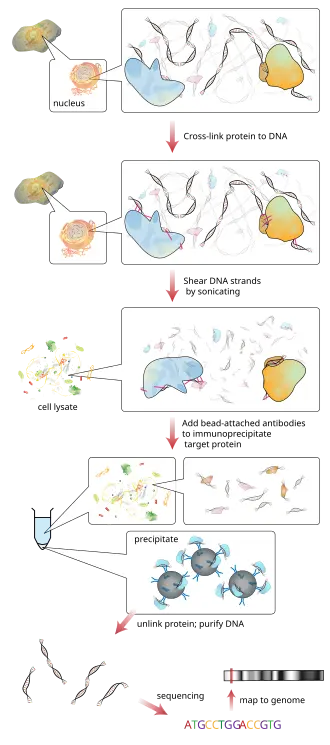
- Chromatin immunoprecipitation (ChIP)
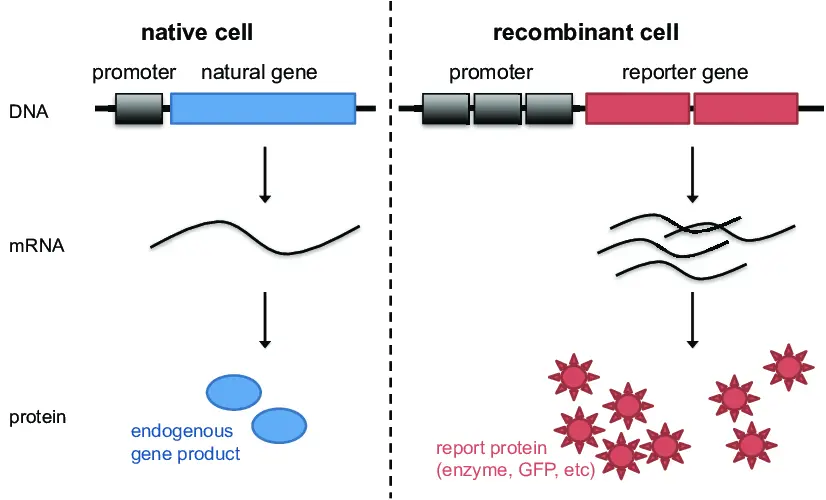
- Reporter gene assays
are essential. These techniques require high-quality, reliable reagents such as:
- Specific antibodies against transcription factors
- DNA probes and oligonucleotides for motif regions
- Enzymes and buffers optimized for DNA-protein interaction studies
Investing in validated reagents ensures reproducibility and robust data, crucial for publishing and downstream applications.
Troubleshooting Tips for Motif Validation Experiments
- Optimize antibody specificity: Use well-characterized antibodies to avoid non-specific binding.
- Design precise probes: Ensure oligonucleotides cover predicted motif sequences without additional flanking regions.
- Maintain proper controls: Include negative controls and competition assays to validate specificity.
- Optimize reaction conditions: Buffer composition, temperature, and incubation times can drastically affect assay sensitivity.
Proper planning and high-quality reagents significantly reduce experimental variability and increase success rates.
Integrating cisRED Data with Your Laboratory Workflow
cisRED data can be seamlessly incorporated into broader experimental designs:
- Select candidate motifs for validation based on conservation and functional annotation.
- Use motif information to design primers and probes for PCR and EMSA.
- Combine with gene expression data to prioritize regulatory elements linked to biological conditions.
This integrated approach accelerates discovery, reduces trial-and-error, and ultimately helps produce more impactful scientific findings.
Conclusion
cisRED is a powerful platform that empowers researchers by providing reliable predictions of conserved cis-regulatory elements, a critical foundation for understanding gene regulation. When paired with the right experimental tools and reagents, these computational insights become actionable hypotheses ready for biological validation.
Whether you’re unraveling fundamental gene regulatory networks or investigating disease-associated regulatory mutations, leveraging cisRED data alongside quality reagents will elevate your research and streamline your path from discovery to publication.